Upload Data From Any Software Pipeline
Limelight is designed to support the native results from any bottom up proteomics pipeline. Results are converted to limelight XML using a converter and any data represented as limelight XML may be uploaded into limelight. Converters currently exist for Comet, Crux, Magnum, MetaMorpheus, MSFragger, Trans-Proteomic Pipeline (TPP), and more. To learn more about converting and uploading results, see limelights’s upload documentation.
All Underlying Data Are Available
All PSM- and peptide-level statistics are available on every page, including the ability to view underlying spectra. Data may be filtered using any of the native scores of whichever search program generated the data.
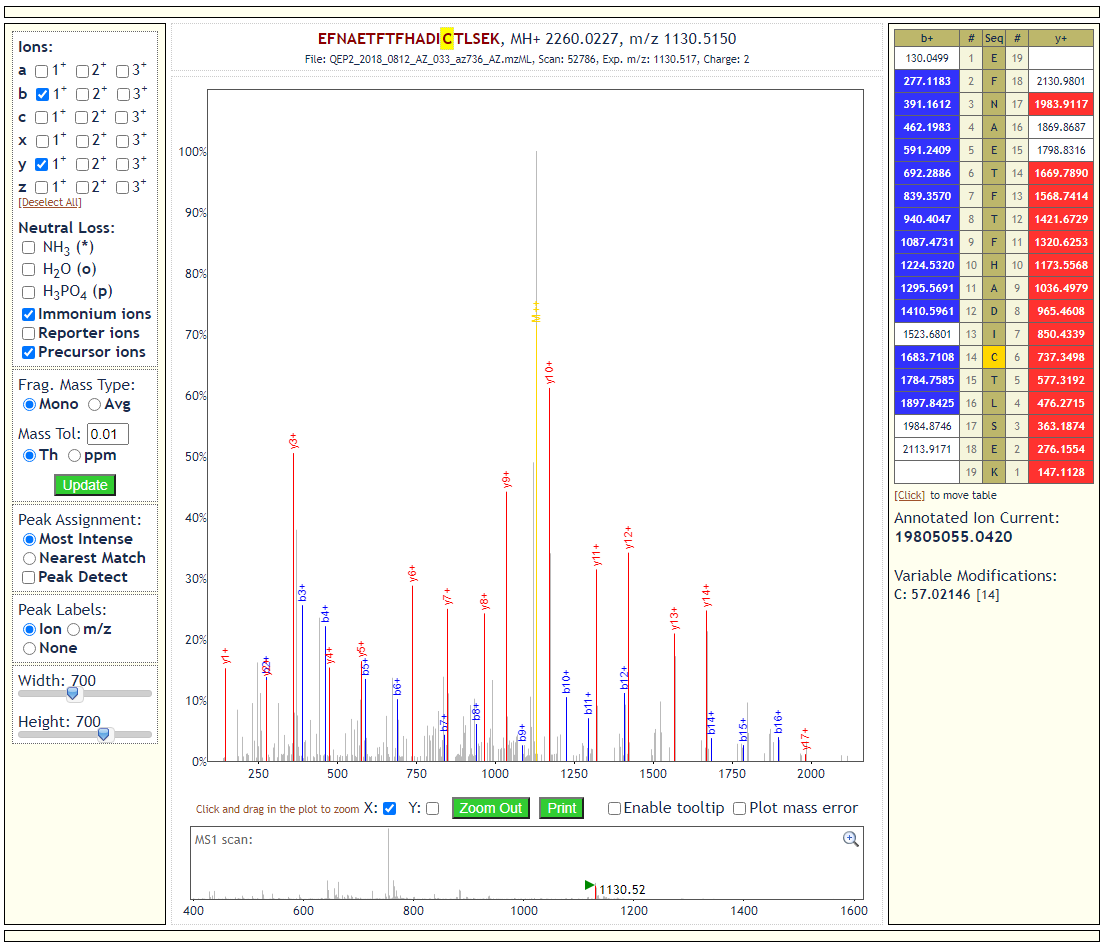
Merge, Compare, and Contrast Data Sets
Limelight allows you to combine, compare, and contrast data from different searches–even if those searches used different search programs (such as Magnum and MSFragger) or different versions (or settings) of the same program. Each combined data set displays and is filtered by native scores from the respective search programs.
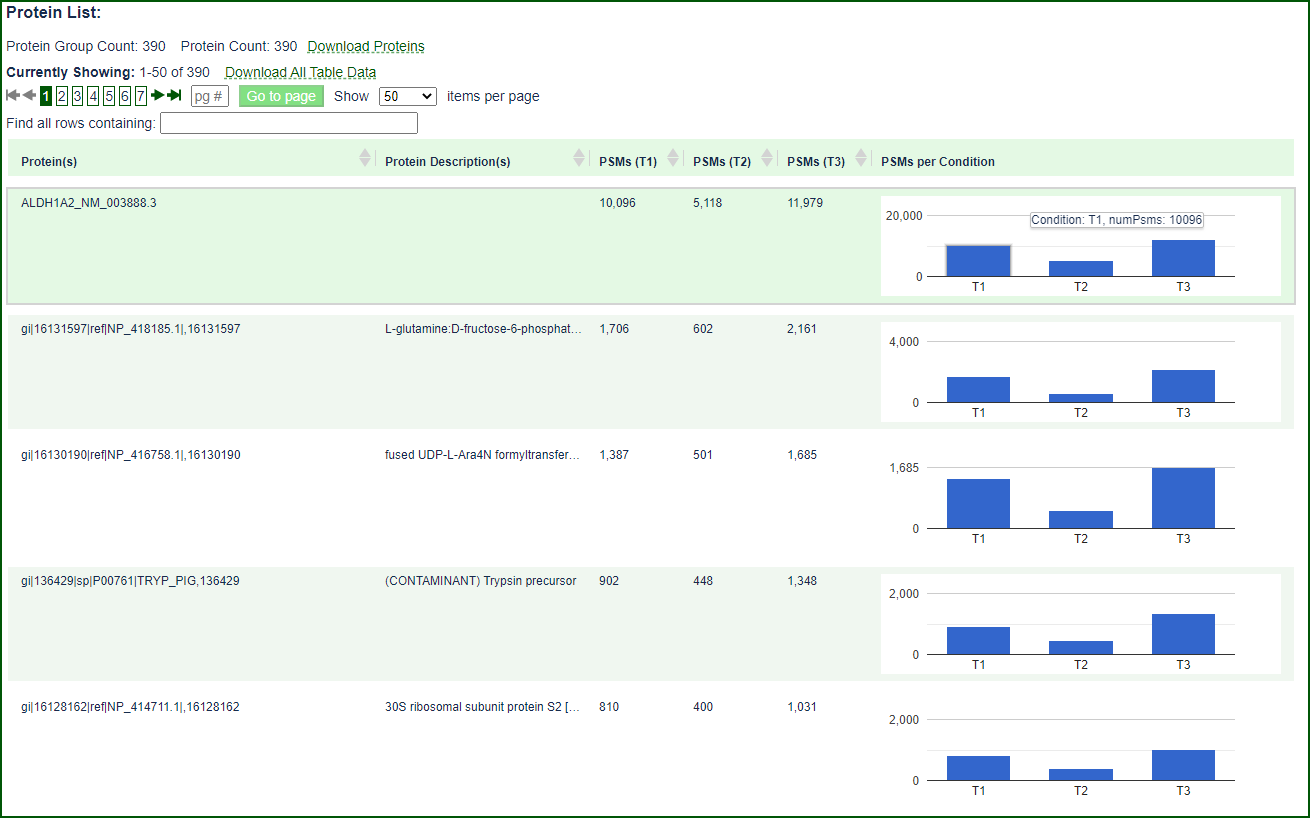
Interactive Analysis Tools and Data Visualization
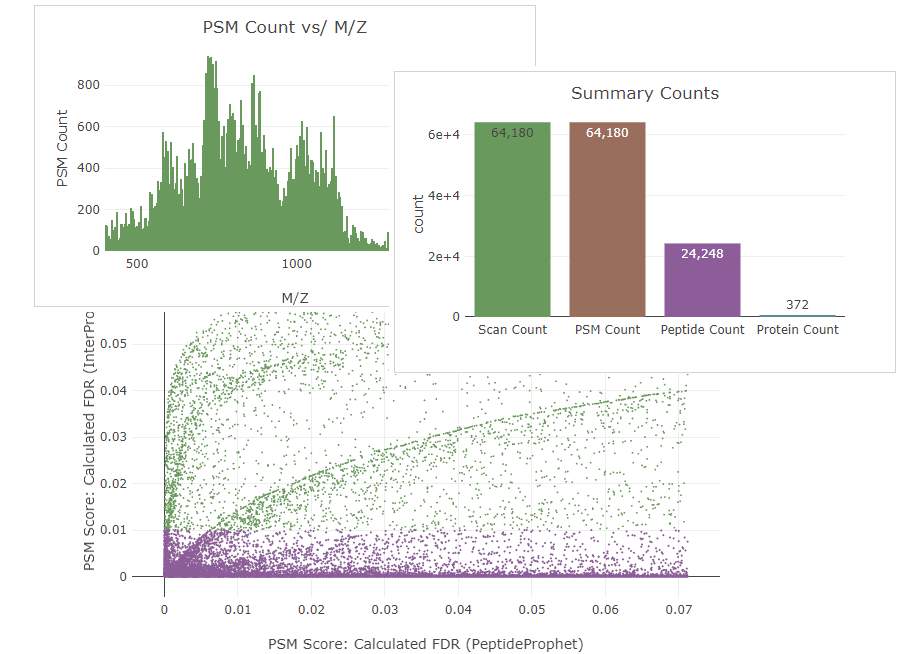
Limelight contains numerous, interactive views of the data–including tables, downloadable reports, and interactive graphical visualizations.
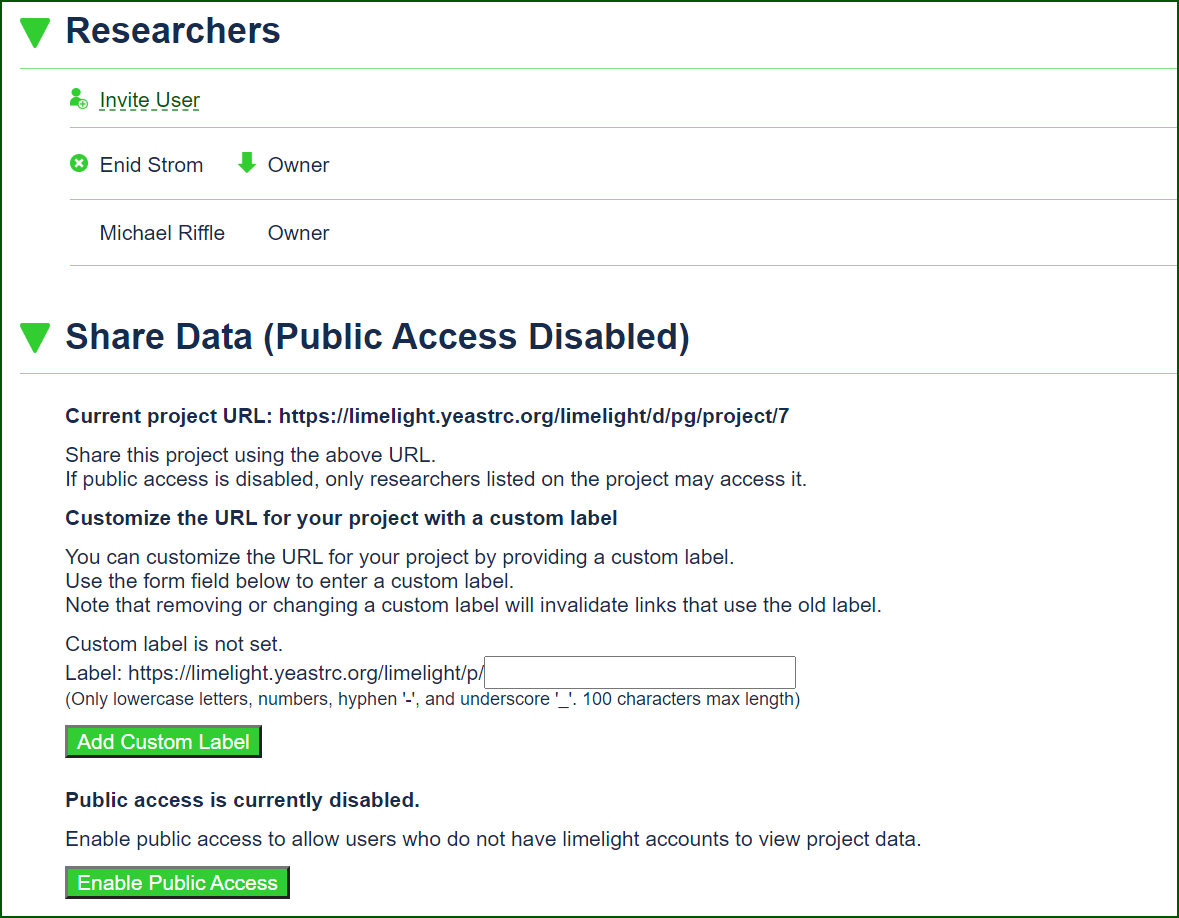
Access Control and Advanced Sharing Tools
Limelight includes sharing tools that make it ideal for collaboration or publication. Access to data is simply and securely controlled by project owners, who can share data by inviting specific users to their project, sharing secure links, or by enabling public access.
Advanced Filtering Options
Limelight includes advanced filtering options to help you focus on the data that matter to you. Filter peptides by pipeline scores, sequence, their uniqueness, protein positions, modification masses, and more. When applicable, all filters use the native scores present in any respective pipeline.
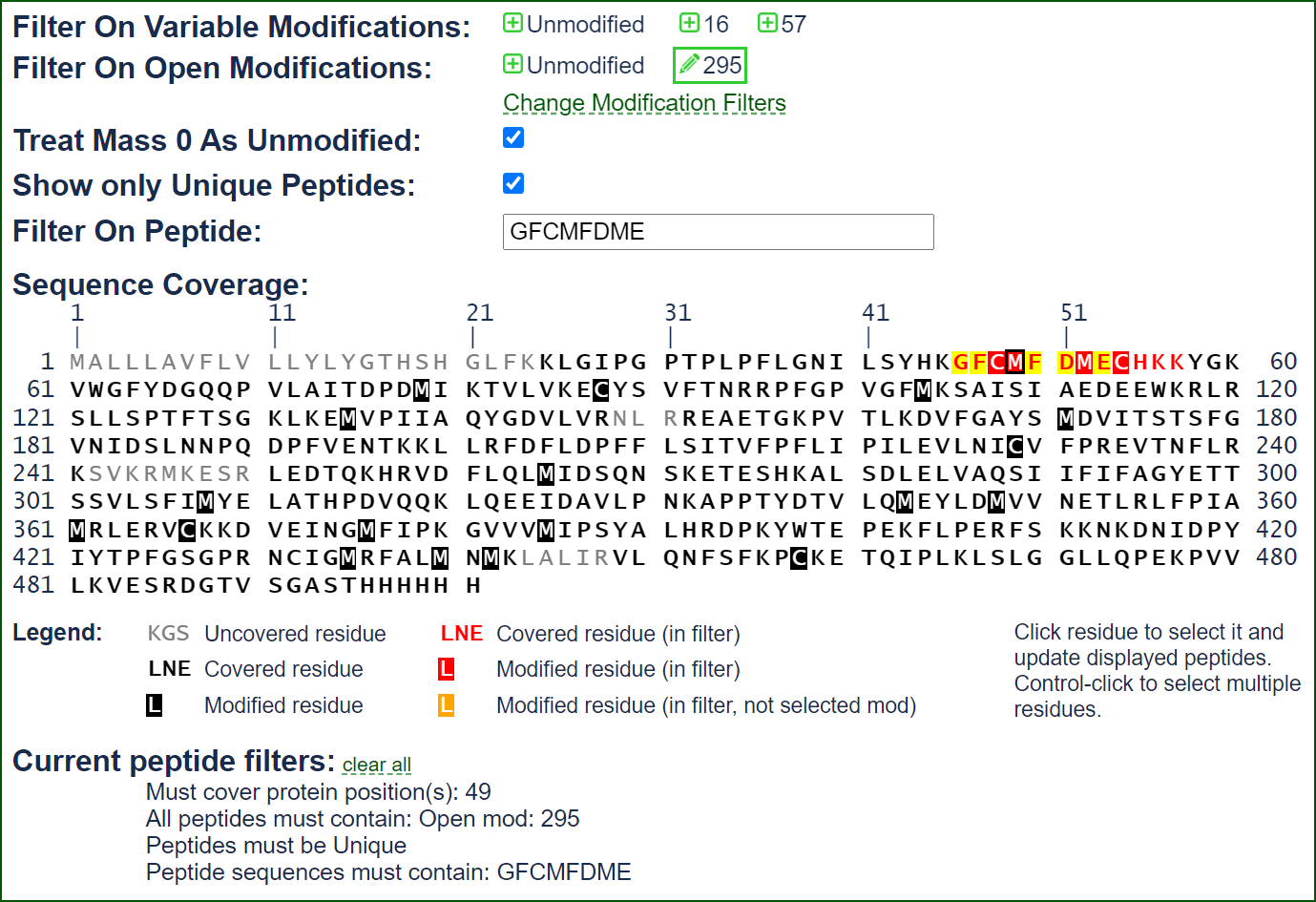
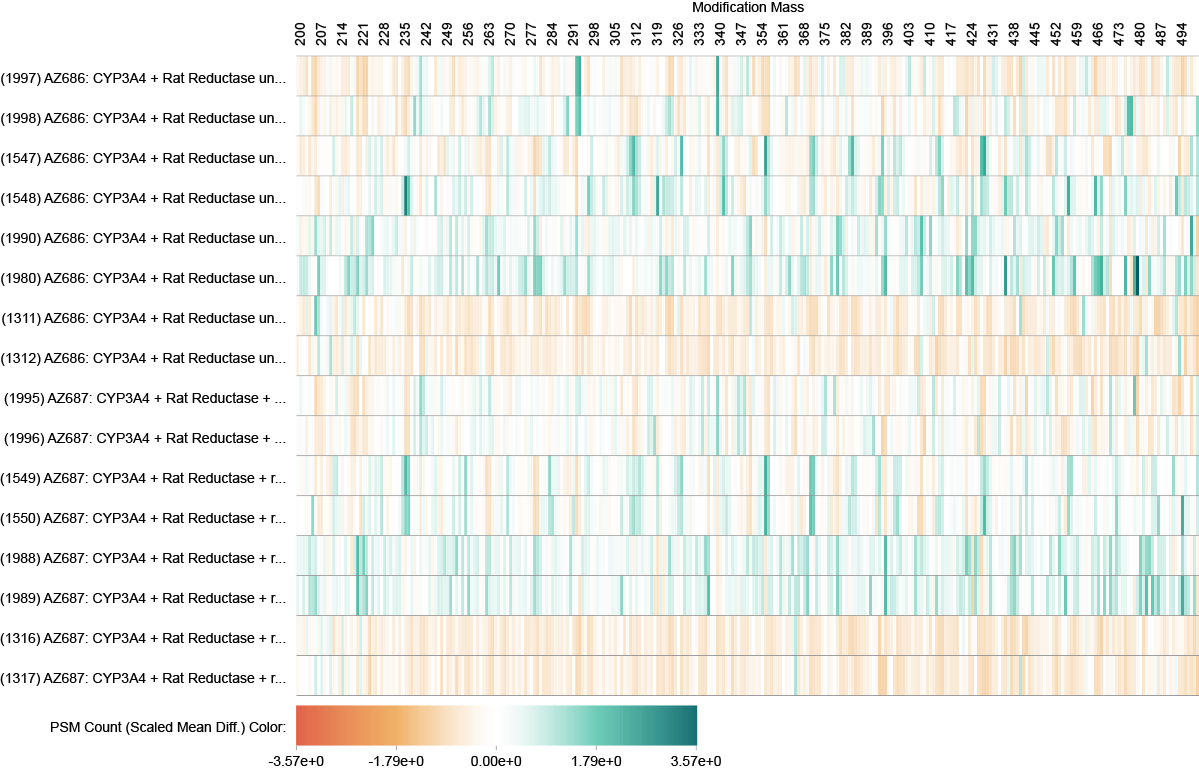
Protein Adduct and Modification Analysis
Limelight has tools specifically for finding protein adducts or post-translational modifications of interest. Visualize how modification masses are enriched or depleted across groups of searches, view reports of which proteins (and positions) are modified with which modification masses, perform statistical analysis, and more.
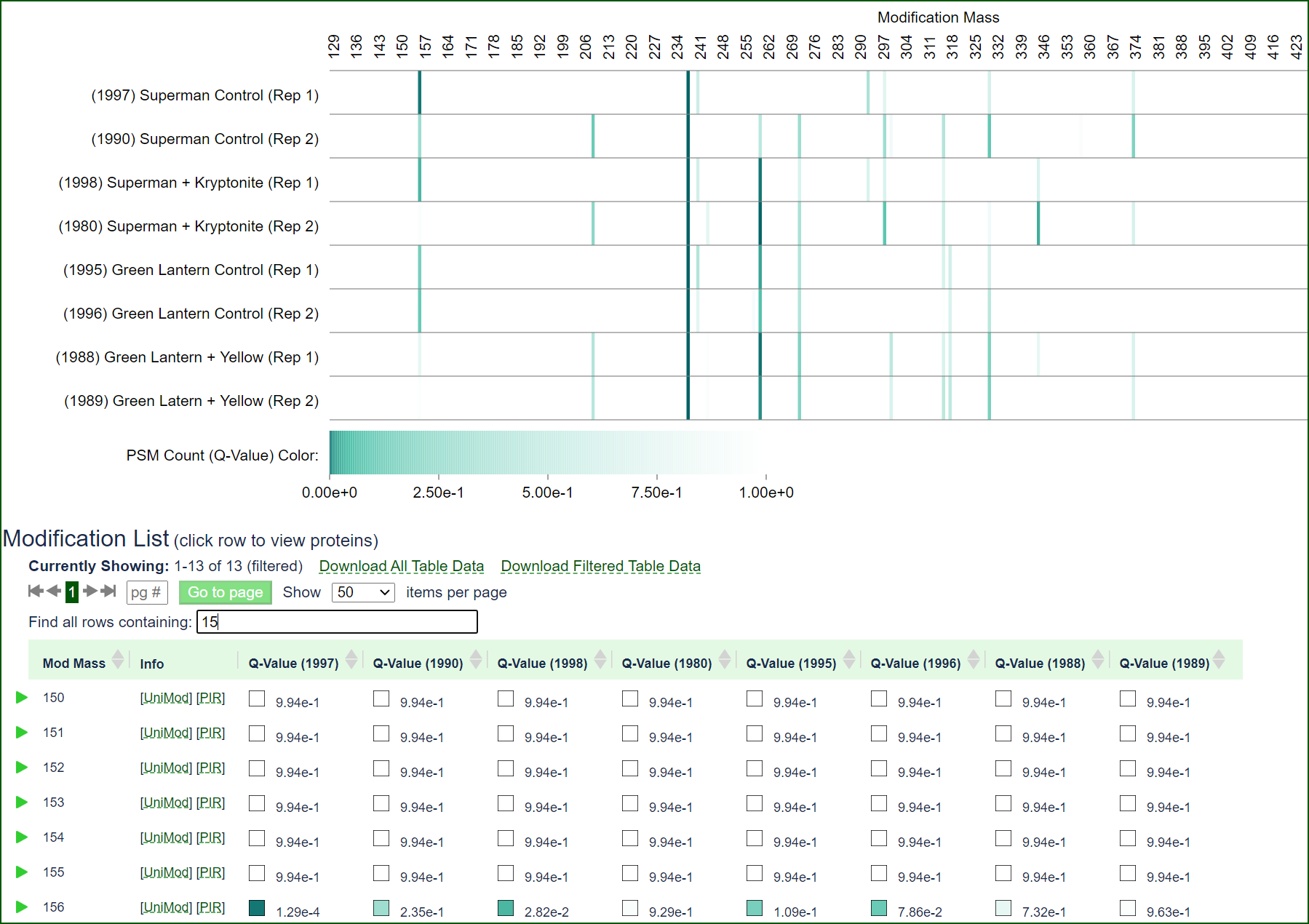
Quality Control (QC) Tools
Limelight contains summary statistics and quality control (QC) tools to compare different searches and find and diagnose any issues.